PUBLICATIONS
RNA exosome drives early B cell development via noncoding RNA processing mechanisms
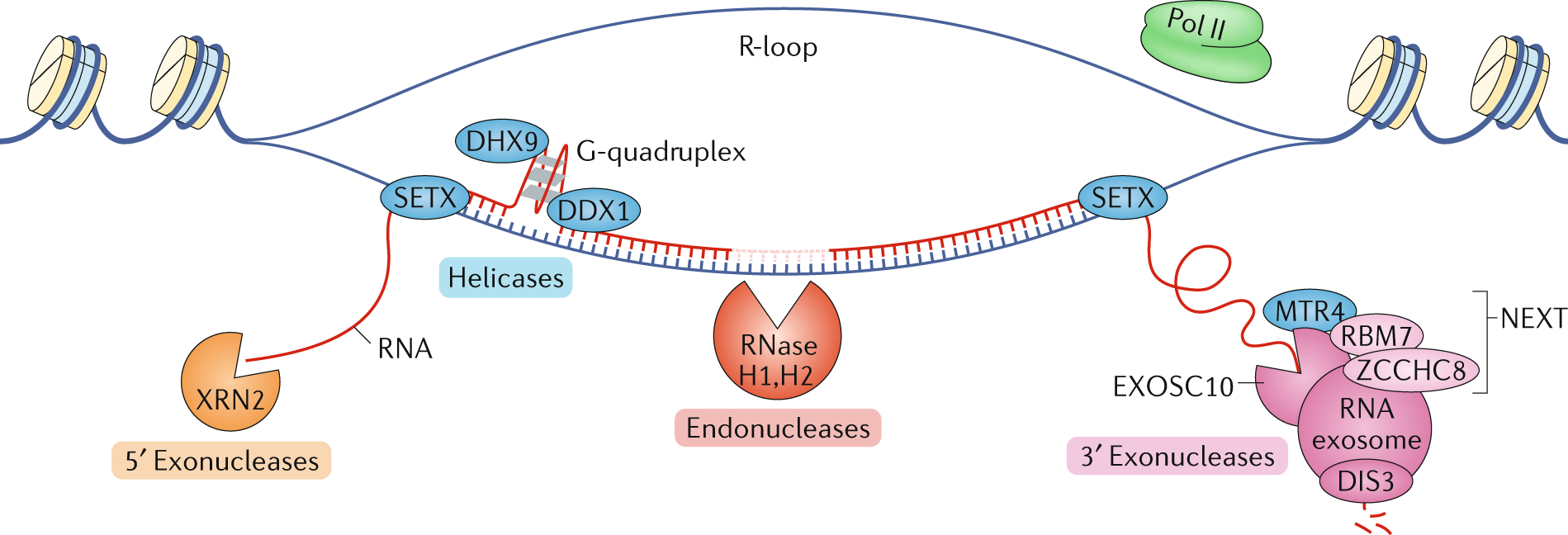
Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery
SELECTED PUBLICATIONS:
Leeman-Neill RJ, Song D, Bizarro J, Wacheul L, Rothschild G, Singh S, Yang Y, Sarode AY, Gollapalli K, Wu L, Zhang W, Chen Y, Lauring MC, Whisenant DE, Bhavsar S, Lim J, Swerdlow SH, Bhagat G, Zhao Q, Berchowitz LE, Lafontaine DLJ, Wang J, Basu U. Noncoding mutations cause super-enhancer retargeting resulting in protein synthesis dysregulation during B cell lymphoma progression. Nat Genet. 2023 Dec;55(12):2160-2174. doi: 10.1038/s41588-023-01561-1. Epub 2023 Dec 4. PMID: 38049665; PMCID: PMC10703697.
Laffleur B, Batista CR, Zhang W, Lim J, Yang B, Rossille D, Wu L, Estrella J, Rothschild G, Pefanis E, Basu U. RNA exosome drives early B cell development via noncoding RNA processing mechanisms. Sci Immunol. 2022 Jun 3;7(72):eabn2738. doi: 10.1126/sciimmunol.abn2738. Epub 2022 Jun 3. PMID: 35658015.
Lekha Nair, Wanwei Zhang, Brice Laffleur, Mukesh K. Jha, Junghyun Lim, Heather Lee, Lijing Wu, Nehemiah S. Alvarez, Zhi-ping Liu, Emilia L. Munteanu, Theresa Swayne, Jacob H. Hanna, Lei Ding, Gerson Rothschild, Uttiya Basu. Mechanism of noncoding RNA-associated N6-methyladenosine recognition by an RNA processing complex during IgH DNA recombination. Molecular Cell. 2021 Aug 20;S1097-2765(21)00615-8. doi.org/10.1016/j.molcel.2021.07.037. PMID: 34450044.
Laffleur B, Lim J, Zhang W, Chen Y, Pefanis E, Bizarro J, Batista CR, Wu L, Economides AN, Wang J, Basu U. Noncoding RNA processing by DIS3 regulates chromosomal architecture and somatic hypermutation in B cells. Nature Genetics. 2021 Feb;53(2):230-242. doi: 10.1038/s41588-020-00772-0. PMID: 33526923.
Rothschild G, Zhang W, Lim J, Giri PK, Laffleur B, Chen Y, Fang M, Chen Y, Nair L, Liu ZP, Deng H, Hammarström L, Wang J, Basu U. Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class switch recombination. Sci Immunol. 2020 Feb 7;5(44):eaay5864. doi: 10.1126/sciimmunol.aay5864. PMID: 32034089.
Lim J, Giri PK, Kazadi D, Laffleur B, Zhang W, Grinstein V, Pefanis E, Brown LM, Ladewig E, Martin O, Chen Y, Rabadan R, Boyer F, Rothschild G, Cogné M, Pinaud E, Deng H, Basu U. Nuclear Proximity of Mtr4 to RNA Exosome Restricts DNA Mutational Asymmetry. Cell. 2017 Apr 20;169(3):523-537.e15. doi: 10.1016/j.cell.2017.03.043. PMID: 28431250; PMCID: PMC5515252.
Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015 May 7;161(4):774-89. doi: 10.1016/j.cell.2015.04.034. PMID: 25957685; PMCID: PMC4428671.
Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014 Oct 16;514(7522):389-93. doi: 10.1038/nature13580. Epub 2014 Aug 6. PMID: 25119026; PMCID: PMC4372240.
Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011 Feb 4;144(3):353-63. doi: 10.1016/j.cell.2011.01.001. Epub 2011 Jan 20. PMID: 21255825; PMCID: PMC3065114.
___
SELECTED REVIEW ARTICLE:
Lauring MC, Basu U. Somatic hypermutation mechanisms during lymphomagenesis and transformation. Curr Opin Genet Dev. 2024 Apr;85:102165. doi: 10.1016/j.gde.2024.102165. Epub 2024 Feb 29. PMID: 38428317.
Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol. 2020 Mar;21(3):123-136. doi: 10.1038/s41580-019-0209-0. Epub 2020 Feb 4. PMID: 32020081; PMCID: PMC7107043.
___
ALL PUBLICATIONS:
Leeman-Neill RJ, Song D, Bizarro J, Wacheul L, Rothschild G, Singh S, Yang Y, Sarode AY, Gollapalli K, Wu L, Zhang W, Chen Y, Lauring MC, Whisenant DE, Bhavsar S, Lim J, Swerdlow SH, Bhagat G, Zhao Q, Berchowitz LE, Lafontaine DLJ, Wang J, Basu U. Noncoding mutations cause super-enhancer retargeting resulting in protein synthesis dysregulation during B cell lymphoma progression. Nat Genet. 2023 Dec;55(12):2160-2174. doi: 10.1038/s41588-023-01561-1. Epub 2023 Dec 4. PMID: 38049665; PMCID: PMC10703697.
Zhang Y, Zhang W, Zhao J, Ito T, Jin J, Aparicio AO, Zhou J, Guichard V, Fang Y, Que J, Urban JF Jr, Hanna JH, Ghosh S, Wu X, Ding L, Basu U, Huang Y. m6A RNA modification regulates innate lymphoid cell responses in a lineage-specific manner. Nat Immunol. 2023 Aug;24(8):1256-1264. doi: 10.1038/s41590-023-01548-4. Epub 2023 Jul 3. PMID: 37400674; PMCID: PMC10797530.
Laffleur B, Batista CR, Zhang W, Lim J, Yang B, Rossille D, Wu L, Estrella J, Rothschild G, Pefanis E, Basu U. RNA exosome drives early B cell development via noncoding RNA processing mechanisms. Sci Immunol. 2022 Jun 3;7(72):eabn2738. doi: 10.1126/sciimmunol.abn2738. Epub 2022 Jun 3. PMID: 35658015.
Lekha Nair, Wanwei Zhang, Brice Laffleur, Mukesh K. Jha, Junghyun Lim, Heather Lee, Lijing Wu, Nehemiah S. Alvarez, Zhi-ping Liu, Emilia L. Munteanu, Theresa Swayne, Jacob H. Hanna, Lei Ding, Gerson Rothschild, Uttiya Basu. Mechanism of noncoding RNA-associated N6-methyladenosine recognition by an RNA processing complex during IgH DNA recombination. Molecular Cell. 2021 Aug 20;S1097-2765(21)00615-8. doi.org/10.1016/j.molcel.2021.07.037. PMID: 34450044.
Laffleur B, Lim J, Zhang W, Chen Y, Pefanis E, Bizarro J, Batista CR, Wu L, Economides AN, Wang J, Basu U. Noncoding RNA processing by DIS3 regulates chromosomal architecture and somatic hypermutation in B cells. Nature Genetics. 2021 Feb;53(2):230-242. doi: 10.1038/s41588-020-00772-0. PMID: 33526923
Rothschild G, Zhang W, Lim J, Giri PK, Laffleur B, Chen Y, Fang M, Chen Y, Nair L, Liu ZP, Deng H, Hammarström L, Wang J, Basu U. Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class switch recombination. Sci Immunol. 2020 Feb 7;5(44):eaay5864. doi: 10.1126/sciimmunol.aay5864. PMID: 32034089.
Lai W.K.M., Mariani L., Rothschild G., Smith E.R., Venters B.J., Blanda T.R., Kuntala P.K., Bocklund K., Mairose J., Dweikat S., Mistretta K., Rossi M. J., James D., Anderson J. T., Phanor S.K., Zhang W., Shaw A., Novitzky K., McAnarney E., Keogh M.C., Shilatifard A,, Basu U., Bulyk M.L., Pugh B.F. (2020) Screening of PCRP transcription factor antibodies in biochemical assays. BIORVIX: https://www.biorxiv.org/content/10.1101/2020.06.08.140046v1
Kane JR, Zhao J, Tsujiuchi T, Laffleur B, Arrieta VA, Mahajan A, Rao G, Mela A, Dmello C, Chen L, Zhang DY, González-Buendia E, Lee-Chang C, Xiao T, Rothschild G, Basu U, Horbinski C, Lesniak MS, Heimberger AB, Rabadan R, Canoll P, Sonabend AM. CD8+ T-cell-Mediated Immunoediting Influences Genomic Evolution and Immune Evasion in Murine Gliomas. Clin Cancer Res. (2020) Aug 15;26(16):4390-4401. doi: 10.1158/1078-0432.CCR-19-3104. Epub 2020 May 19. PMID: 32430477; PMCID: PMC7442699.
Sun J., Basu U. (2020) Purification of murine IL10+ B cells for analyses of biological function and transcriptomics. Methods in Molecular Biology (in press).
Kazadi D, Lim J, Rothschild G, Grinstein V, Laffleur B, Becherel O, Lavin MJ, Basu U. Effects of senataxin and RNA exosome on B-cell chromosomal integrity. Heliyon. 2020 Mar 12;6(3):e03442. doi: 10.1016/j.heliyon.2020.e03442. PMID: 32195383; PMCID: PMC7075999.
Laffleur B, Basu U. Biology of RNA Surveillance in Development and Disease. Trends Cell Biol. 2019 May;29(5):428-445. doi: 10.1016/j.tcb.2019.01.004. Epub 2019 Feb 10. PMID: 30755352; PMCID: PMC6581522.
Rothschild G, Basu U. Lingering Questions about Enhancer RNA and Enhancer Transcription-Coupled Genomic Instability. Trends Genet. 2017 Feb;33(2):143-154. doi: 10.1016/j.tig.2016.12.002. Epub 2017 Jan 10. PMID: 28087167; PMCID: PMC5291171.
Skamagki M, Zhang C, Ross CA, Ananthanarayanan A, Liu Z, Mu Q, Basu U, Wang J, Zhao R, Li H, Kim K. RNA Exosome Complex-Mediated Control of Redox Status in Pluripotent Stem Cells. Stem Cell Reports. 2017 Oct 10;9(4):1053-1061. doi: 10.1016/j.stemcr.2017.08.024. PMID: 29020613; PMCID: PMC5639470.
Lim J, Giri PK, Kazadi D, Laffleur B, Zhang W, Grinstein V, Pefanis E, Brown LM, Ladewig E, Martin O, Chen Y, Rabadan R, Boyer F, Rothschild G, Cogné M, Pinaud E, Deng H, Basu U. Nuclear Proximity of Mtr4 to RNA Exosome Restricts DNA Mutational Asymmetry. Cell. 2017 Apr 20;169(3):523-537.e15. doi: 10.1016/j.cell.2017.03.043. PMID: 28431250; PMCID: PMC5515252.
Rialdi A, Hultquist J, Jimenez-Morales D, Peralta Z, Campisi L, Fenouil R, Moshkina N, Wang ZZ, Laffleur B, Kaake RM, McGregor MJ, Haas K, Pefanis E, Albrecht RA, Pache L, Chanda S, Jen J, Ochando J, Byun M, Basu U, García-Sastre A, Krogan N, van Bakel H, Marazzi I. The RNA Exosome Syncs IAV-RNAPII Transcription to Promote Viral Ribogenesis and Infectivity. Cell. 2017 May 4;169(4):679-692.e14. doi: 10.1016/j.cell.2017.04.021. PMID: 28475896; PMCID: PMC6217988.
Sun J, Wang J, Pefanis E, Chao J, Rothschild G, Tachibana I, Chen JK, Ivanov II, Rabadan R, Takeda Y, Basu U. Transcriptomics Identify CD9 as a Marker of Murine IL-10-Competent Regulatory B Cells. Cell Rep. 2015 Nov 10;13(6):1110-1117. doi: 10.1016/j.celrep.2015.09.070. Epub 2015 Oct 29. PMID: 26527007; PMCID: PMC4644501.
Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015 May 7;161(4):774-89. doi: 10.1016/j.cell.2015.04.034. PMID: 25957685; PMCID: PMC4428671.
Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014 Oct 16;514(7522):389-93. doi: 10.1038/nature13580. Epub 2014 Aug 6. PMID: 25119026; PMCID: PMC4372240.
Sun J, Keim CD, Wang J, Kazadi D, Oliver PM, Rabadan R, Basu U. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes Dev. 2013 Aug 15;27(16):1821-33. doi: 10.1101/gad.210211.112. PMID: 23964096; PMCID: PMC3759698.
Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011 Jun;12(6):536-43. doi: 10.1038/ni.2037. Epub 2011 May 15. PMID: 21572431; PMCID: PMC3117275.
Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011 Feb 4;144(3):353-63. doi: 10.1016/j.cell.2011.01.001. Epub 2011 Jan 20. PMID: 21255825; PMCID: PMC3065114.
Keim C, Grinstein V, Basu U. Recombinant retroviral production and infection of B cells. J Vis Exp. 2011 Feb 18;(48):2371. doi: 10.3791/2371. PMID: 21372789; PMCID: PMC3197389.
Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, Chaudhuri J. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci U S A. 2009 Feb 24;106(8):2717-22. doi: 10.1073/pnas.0812304106. Epub 2009 Feb 5. Erratum in: Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5731. PMID: 19196992; PMCID: PMC2650332.
Basu U, Wang Y, Alt FW. Evolution of phosphorylation-dependent regulation of activation-induced cytidine deaminase. Mol Cell. 2008 Oct 24;32(2):285-91. doi: 10.1016/j.molcel.2008.08. PMID: 18951095; PMCID: PMC2597080.
Ray P, Basu U, Ray A, Majumdar R, Deng H, Maitra U. The Saccharomyces cerevisiae 60 S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem. 2008 Apr 11;283(15):9681-91. doi: 10.1074/jbc.M710294200. Epub 2008 Feb 5. PMID: 18256024; PMCID: PMC2442299.
Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005 Nov 24;438(7067):508-11. doi: 10.1038/nature04255. Epub 2005 Oct 26. PMID: 16251902.
Basu U, Si K, Deng H, Maitra U. Phosphorylation of mammalian eukaryotic translation initiation factor 6 and its Saccharomyces cerevisiae homologue Tif6p: evidence that phosphorylation of Tif6p regulates its nucleocytoplasmic distribution and is required for yeast cell growth. Mol Cell Biol. 2003 Sep;23(17):6187-99. doi: 10.1128/mcb.23.17.6187-6199.2003. PMID: 12917340; PMCID: PMC180954.
Basu U, Si K, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol. 2001 Mar;21(5):1453-62. doi: 10.1128/MCB.21.5.1453-1462.2001. PMID: 11238882; PMCID: PMC86691.
___
REVIEW ARTICLES:
Lauring MC, Basu U. Somatic hypermutation mechanisms during lymphomagenesis and transformation. Curr Opin Genet Dev. 2024 Apr;85:102165. doi: 10.1016/j.gde.2024.102165. Epub 2024 Feb 29. PMID: 38428317.
Nair L., Chung H., and Basu U. (2020) Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nature Reviews Molecular Biology (3): 123-136.
Laffleur B., Basu U. (2019) Biology of RNA surveillance in development and disease. Trends Cell Biol. 29(5): 428-445.
Leeman-Neill R.J., Lim, J., and Basu, U. (2018) The common key to class-switch recombination and somatic hypermutation: Discovery of AID and its role in antibody gene diversification. The Journal of Immunology 201(9): 2527-2529.
Zou, F., Wang, X., Han, X., Rothschild, G., Zheng, S.G., Basu, U., Sun, J. (2018) Expression and Function of Tetraspanins and their Interacting Partners in B cells. Frontiers in Immunology 9. Article 1606 (pp 1-17).
Rothschild, G., and Basu, U. (2017) Lingering questions about enhancer RNA and enhancer transcription-coupled genomic instability. Trends in Genetics 33(2):143-154.
Laffleur, B., Basu, U., Lim, J. (2017) RNA exosome and non-coding RNA-coupled mechanisms in AID-mediated genomic instability. Journal of Molecular Biology 429(21) 3230-3241.
Cassellas R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., Di Noia J.M. (2016) Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nature Reviews Immunology 16(03):164-176.
Rothschild G., Von Krusenstiern A.N., Basu U. (2015) Malaria-induced B cell Genomic Instability, Cell, 162(4): 697-8. Pefanis E. and Basu U. (2015) RNA exosome regulates AID DNA mutator activity in the B cell genome. Advances in Immunology 127: 257-308.
Chao, J., Rothschild, G., Basu, U. (2014) Ubiquitination events that regulate recombination of immunoglobulin loci gene segments. Front. Immunol. 5:100, 1-12.
Sun J., Rothschild G., Pefanis E., Basu U. (2013) Transcriptional stalling in B-lymphocytes: A mechanism for antibody diversification and maintenance of genomic integrity. Transcription 4(3):1-9.
Keim, C., Kazadi, D., Rothschild, G., Basu, U. (2013) Regulation of AID, the B cell genome mutator. Genes and Development 27(1):1-17.
Basu, U., Franklin, A., Schwer, B., Cheng, H-L, Chaudhuri, J., Alt, F.W. (2009) Regulation of Activation-Induced Cytidine Deaminase DNA Deamination Activity in B cells by Serine-38 phosphorylation. Biochem Soc Trans.37(Pt 3):561-8.
Basu, U., Franklin, A. and Alt, F.W. (2009) Post-translational regulation of activation induced deaminase. Philosophical Transactions of the Royal Society of Sciences 364(1517):667-73.
Chaudhuri, J., Basu, U., Zarrin, A., Yan, C., Franco, S., Perlot, T., Vuong, B., Wang, J., Phan, R.T., Datta, A., Manis, J., and Alt, F.W. (2007) Evolution of the Immunoglobulin Heavy Chain Class Switch Recombination Mechanism. Advances in Immunology 94:157-214.
Longerich, S., Basu, U., Alt, F.W., Storb U. (2006) AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 18(2), 164-174.
Basu, U., Chaudhuri, J., Phan, R.T., Datta, A., and Alt, F.W. (2007) Regulation of activation induced deaminase via phosphorylation. In: Mechanisms of Lymphocyte Activation and Immune Regulation XI, Gupta, S., Cooper, M., Rajewsky, K., Alt, F.W., Melchers, F., eds. New York, Springer. pp. 129-137.
Basu U. (2004) Eukaryotic Translation Initiation Factor 6 (eIF6) and Regulation of 60S Ribosome Biogenesis, Ph.D. Dissertation, Albert Einstein College of Medicine, Yeshiva University.
___